BenchMark™ Microbial Amplicon Sequencing
BenchMark is our optimized and low-bias approach to amplicon sequencing for microbiome profiling.
Every BenchMark project includes a Core Analysis™ Report, which provides fully annotated data tables, interactive visualizations and all the raw data delivered through our customer portal.
Optimized Amplicon Sequencing
Amplicon sequencing uses primers that match the conserved areas of marker genes, such as the 16S ribosomal RNA gene of bacteria (16S rRNA) or the internal transcribed spacer region 2 of fungi (ITS2).
We offer sequencing that targets the variable region 4 (V4) or variable regions 1 through 3 (V1-V3) of the bacterial 16S rRNA gene, the fungal ITS2 region, and custom marker genes of your choice.
We pair our low-bias amplification method with an optimal closed-reference alignment to provide you with data that is more accurate than standard methods.
Eliminate Taxonomic Dropout
In 2016, our approach was published in Nature Biotechnology by our founders, Daryl Gohl, Dan Knights and Kenny Beckman [1].
Our amplicon sequencing combines real-time PCR sample quantification, dual-indexed multiplexing, and optimized enzymatic and PCR conditions to eliminate most of the taxonomic dropout that is observed with standard amplicon sequencing methods.
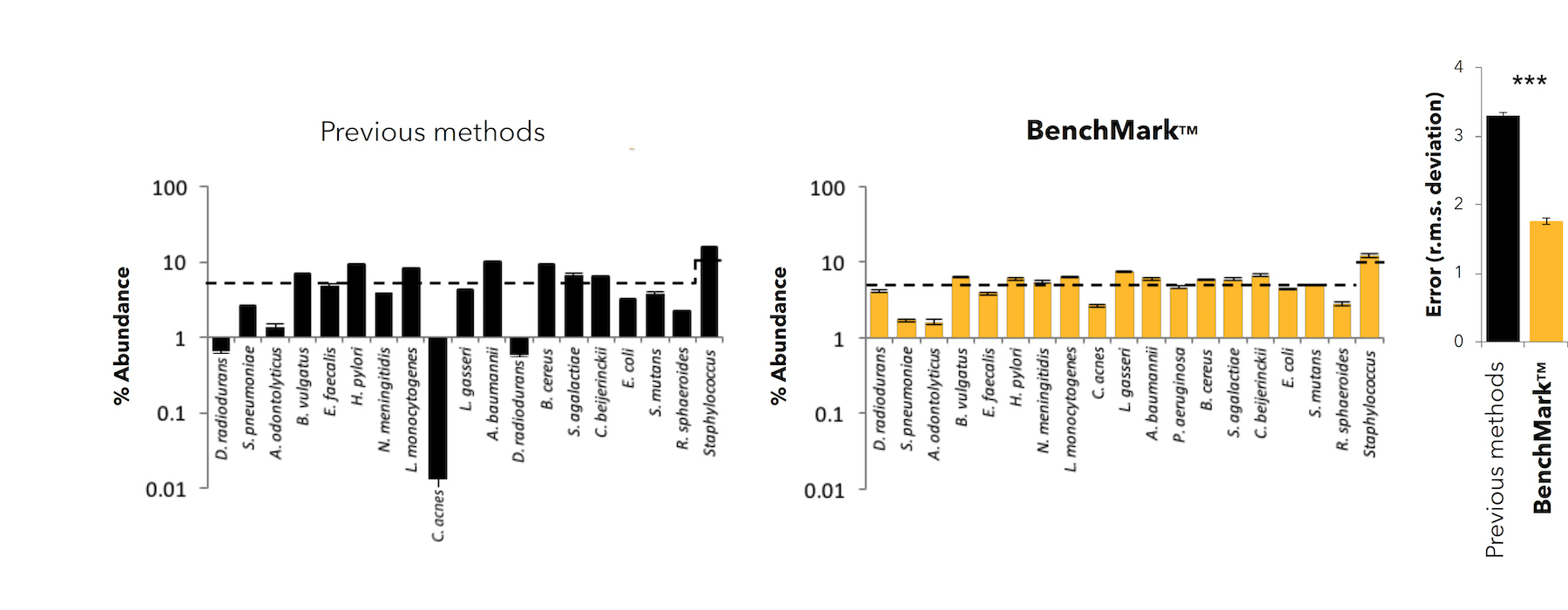
Our approach also suppresses chimera formation and minimizes PCR error, resulting in greater accuracy when tested head-to-head against other amplicon sequencing methods in the analysis of defined mock community standards.
In the plot below, the dotted line represents the expected percent abundance of each community member in a mock community. The BenchMark low-bias approach, highlighted in yellow, comes closest to the expected percent abundance.
Sample Types Suited for BenchMark Amplicon Sequencing
Choosing the right sequencing approach for your research goals is very important.
Our amplicon sequencing approach is perfect for samples that have low biomass, including environmental samples such as soil and surface swabs. We also recommend amplicon sequencing for samples with high amounts of host DNA, such as skin swabs or biopsy samples.
While BenchMark works very well with high biomass samples like stool, depending on your research goals we often recommend BoosterShot®, our Shallow Shotgun approach or DeepSeq™, our whole genome sequencing service for this sample type.
Get in touch if you want to discuss the best sequencing approach for your research goals.
BenchMark Amplicon Sequencing Basics
Suggested Sample Types
BenchMark is great for high biomass samples like stool, but it is particularly useful for samples with low biomass like soil or surface swabs, or samples with high host DNA, like skin or biopsy samples. For more information regarding sample preparation and shipment, check out our sample guidelines.
Diversigen has experience working with a wide array of samples types, some of the most common that are suitable for amplicon sequencing are listed below. Contact us if your sample of interest is not represented here.
- Primary Samples (ex. stool, saliva, biopsies, urine, GI contents)
- Mixed microbial communities and microbial consortia
- Swabs (skin, nasal, vaginal, oral, rectal, environmental/surface)
- Soil and other environmental samples
- pre-extracted DNA (for the best data quality, please allow us to perform the extraction)
Specifications:
We offer sequencing that targets the variable region 4 (V4) or variable regions 1 through 3 (V1-V3) of the bacterial 16S rRNA gene. We also offer amplicon sequencing that targets the fungal ITS2 region or custom marker genes of your choice.
Every BenchMark project includes
- Initial quality control check on all samples
- QC report returned for review
- Amplification and library preparation using our optimized approach
- Core Analysis Report
- Taxonomic assignments via fully optimal gapped DNA sequence alignment to a curated reference database
- Functional predictions (16S only) via alignment to a curated reference database
- Sequencing quality-control dashboard
- Interactive visualizations that leverage your sample metadata
- Fastqs: adaptor-trimmed and quality-filtered sequencing data
- Fasta: trimmed, stitched and combined DNA sequences in QIIME-ready
Need more analytics to support your research goals? Talk to one of our bioinformaticians about our fully-customizable CorePlus Analysis.
Turnaround Times:
Project turnaround times can be adjusted to meet your needs.
In general, our turnaround times are 4-6 weeks depending on the size of your project.
Diversigen provides services and generates data for research purposes only. Not all services are available in all regions.


